This is a lightweight implementation of my pmc package
focusing on what I think are the more common use cases (e.g. it will no
longer support comparisons of a geiger model against an
ouch model). Further, it does not cover many of the newer
model fitting that have been implemented since pmc was
first released.
The goal of this release is mostly to provide compatibility with
current versions of geiger.
Install the package:
library("devtools")
install_github("cboettig/pmc")A trivial example with data simulated from the lambda
model.
library("pmc")
library("geiger")
#> Loading required package: ape
#> Loading required package: phytools
#> Loading required package: maps
library("phytools")
phy <- sim.bdtree(n=10)
dat <- sim.char(rescale(phy, "lambda", .5), 1)[,1,]
out <- pmc(phy, dat, "BM", "lambda", nboot = 50)
#> Warning in geiger::fitContinuous(phy = tree, dat = data, model = model, :
#> Parameter estimates appear at bounds:
#> lambdaPlot the results:
dists <- data.frame(null = out$null, test = out$test)
library("ggplot2")
library("tidyr")
library("dplyr")
#>
#> Attaching package: 'dplyr'
#> The following object is masked from 'package:ape':
#>
#> where
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
dists %>%
gather(dist, value) %>%
ggplot(aes(value, fill = dist)) +
geom_density(alpha = 0.5) +
geom_vline(xintercept = out$lr)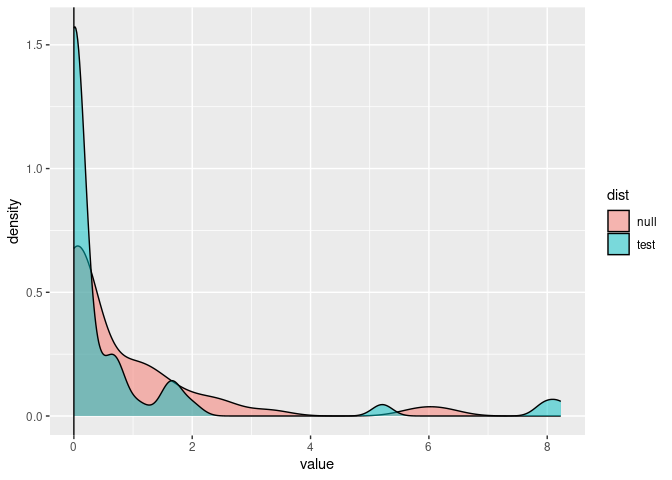
Carl Boettiger, Graham Coop, Peter Ralph (2012) Is your phylogeny informative? Measuring the power of comparative methods, Evolution 66 (7) 2240-51. https://doi.org/10.1111/j.1558-5646.2011.01574.x