CIDER is a meta-clustering workflow designed to handle scRNA-seq data that span multiple samples or conditions. Often, these datasets are confounded by batch effects or other variables. Many existing batch-removal methods assume near-identical cell population compositions across samples. CIDER, in contrast, leverages inter-group similarity measures to guide clustering without requiring such strict assumptions.
You can install CIDER from github with:
# install.packages("devtools")
devtools::install_github('zhiyuan-hu-lab/CIDER')If you have already integrated your scRNA-seq data (e.g., using Seurat-CCA, Harmony, or Scanorama) and want to evaluate how well the biological populations align post-integration, you can use CIDER as follows.
seu.integrated) with corrected PCs
inseu.integrated@reductions$pca@cell.embeddings`seu.integrated@reductions$pca@cell.embeddings <- corrected.PCslibrary(CIDER)
seu.integrated <- hdbscan.seurat(seu.integrated)
ider <- getIDEr(seu.integrated, verbose = FALSE)
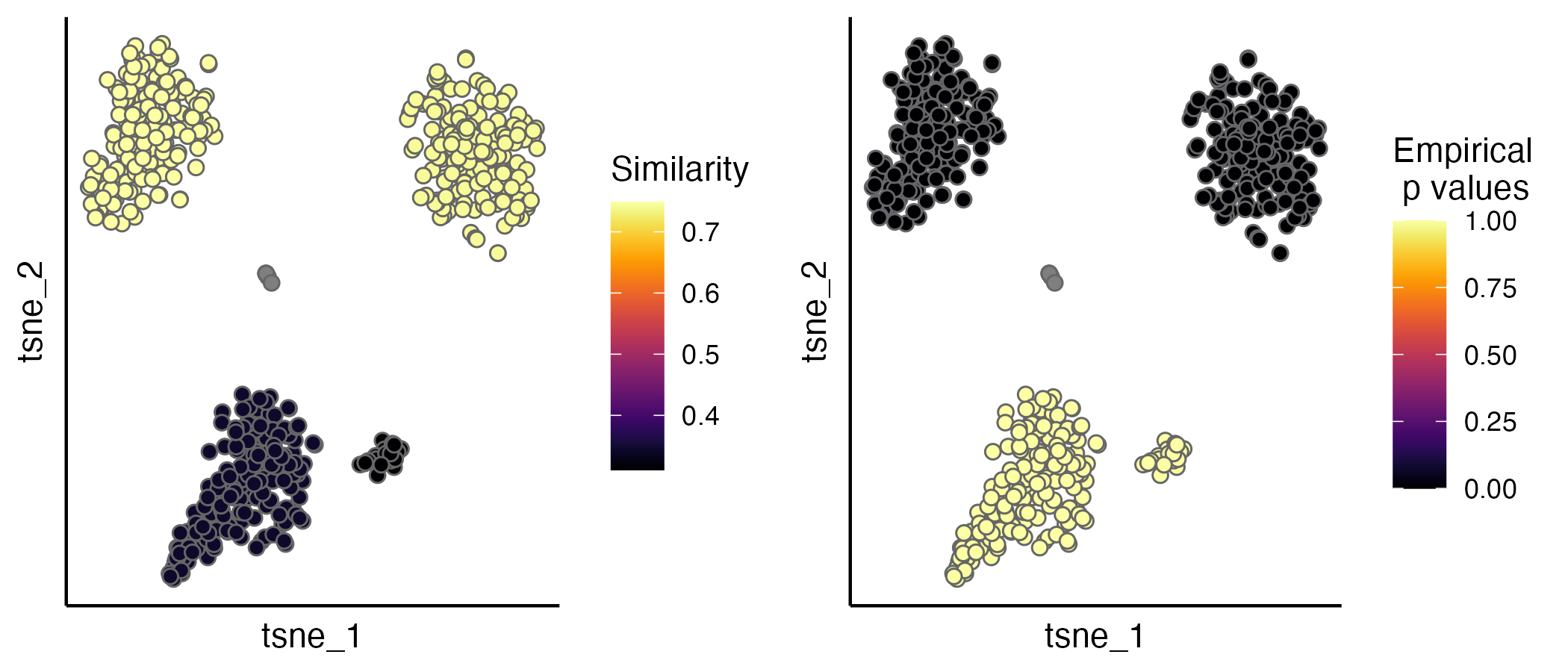
seu.integrated <- estimateProb(seu.integrated, ider)The evaluation scores (IDER-based similarity and empirical p values)
can be visualised by the scatterPlot function.
p1 <- scatterPlot(seu.integrated, "tsne", colour.by = "similarity")
p2 <- scatterPlot(seu.integrated, "tsne", colour.by = "pvalue")
plot_grid(p1,p2, ncol = 2)
For a more detailed walkthrough, see the detailed tutorial of evaluation
In many scenarios, you do not start with an integrated Seurat object but still need to cluster multi-batch scRNA-seq data in a robust way. CIDER provides meta-clustering approaches:
If your Seurat object (seu) has:
initial_cluster in seu@meta.data for
per-batch clusters, andBatch for batch labels,then two main steps are:
# Step 1: Compute IDER-based similarity
ider <- getIDEr(seu,
group.by.var = "initial_cluster",
batch.by.var = "Batch")
# Step 2: Perform final clustering
seu <- finalClustering(seu, ider, cutree.h = 0.45)The final clusters will be stored in
seu@meta.data$final_cluster (by default).
If you find CIDER helpful for your research, please cite:
Z. Hu, A. A. Ahmed, C. Yau. CIDER: an interpretable meta-clustering framework for single-cell RNA-seq data integration and evaluation. Genome Biology 22, Article number: 337 (2021); doi: https://doi.org/10.1186/s13059-021-02561-2
Z. Hu, M. Artibani, A. Alsaadi, N. Wietek, M. Morotti, T. Shi, Z. Zhong, L. Santana Gonzalez, S. El-Sahhar, M. KaramiNejadRanjbar, G. Mallett, Y. Feng, K. Masuda, Y. Zheng, K. Chong, S. Damato, S. Dhar, L. Campo, R. Garruto Campanile, V. Rai, D. Maldonado-Perez, S. Jones, V. Cerundolo, T. Sauka-Spengler, C. Yau, A. A. Ahmed. The repertoire of serous ovarian cancer non-genetic heterogeneity revealed by single-cell sequencing of normal fallopian tube epithelial cells. Cancer Cell 37 (2), p226-242.E7 (2020). doi: https://doi.org/10.1101/2021.03.29.437525