
Julia (mixed-effects) regression modelling from R
You can install the development version of jlme from GitHub with:
# install.packages("devtools")
devtools::install_github("yjunechoe/jlme")library(jlme)
jlme_setup(){jlme} uses {JuliaConnectoR} to connect to
a Julia session. See JuliaConnectoR
package documentation for troubleshooting related to Julia
installation and configuration.
{jlme}Once set up, (g)lm() and (g)lmer
complements in Julia are available via jlm() and
jlmer(), respectively.
jlm() with lm()/glm()
syntax:
# lm(mpg ~ hp, mtcars)
jlm(mpg ~ hp, mtcars)
#> <Julia object of type StatsModels.TableRegressionModel>
#>
#> mpg ~ 1 + hp
#>
#> ────────────────────────────────────────────────────────────────────────────
#> Coef. Std. Error z Pr(>|z|) Lower 95% Upper 95%
#> ────────────────────────────────────────────────────────────────────────────
#> (Intercept) 30.0989 1.63392 18.42 <1e-75 26.8964 33.3013
#> hp -0.0682283 0.0101193 -6.74 <1e-10 -0.0880617 -0.0483948
#> ────────────────────────────────────────────────────────────────────────────Contrasts in factor columns are preserved:
x <- mtcars
# Sum code `am`
x$am_sum <- factor(x$am)
contrasts(x$am_sum) <- contr.sum(2)
# Helmert code `cyl`
x$cyl_helm <- factor(x$cyl)
contrasts(x$cyl_helm) <- contr.helmert(3)
colnames(contrasts(x$cyl_helm)) <- c("4vs6", "4&6vs8")
jlm(mpg ~ am_sum + cyl_helm, x)
#> <Julia object of type StatsModels.TableRegressionModel>
#>
#> mpg ~ 1 + am_sum + cyl_helm
#>
#> ───────────────────────────────────────────────────────────────────────────────
#> Coef. Std. Error z Pr(>|z|) Lower 95% Upper 95%
#> ───────────────────────────────────────────────────────────────────────────────
#> (Intercept) 20.6739 0.572633 36.10 <1e-99 19.5516 21.7963
#> am_sum: 1 -1.27998 0.648789 -1.97 0.0485 -2.55158 -0.00837293
#> cyl_helm: 4vs6 -3.07806 0.767861 -4.01 <1e-04 -4.58304 -1.57308
#> cyl_helm: 4&6vs8 -2.32983 0.414392 -5.62 <1e-07 -3.14203 -1.51764
#> ───────────────────────────────────────────────────────────────────────────────jlmer() with lmer()/glmer()
syntax:
# lme4::lmer(Reaction ~ Days + (Days | Subject), sleepstudy)
jlmer(Reaction ~ Days + (Days | Subject), lme4::sleepstudy, REML = TRUE)
#> <Julia object of type LinearMixedModel>
#>
#> Reaction ~ 1 + Days + (1 + Days | Subject)
#>
#> Variance components:
#> Column Variance Std.Dev. Corr.
#> Subject (Intercept) 612.10016 24.74066
#> Days 35.07171 5.92214 +0.07
#> Residual 654.94001 25.59180
#> ──────────────────────────────────────────────────
#> Coef. Std. Error z Pr(>|z|)
#> ──────────────────────────────────────────────────
#> (Intercept) 251.405 6.8246 36.84 <1e-99
#> Days 10.4673 1.54579 6.77 <1e-10
#> ──────────────────────────────────────────────────# lme4::glmer(r2 ~ Anger + Gender + (1 | id), VerbAgg, family = "binomial")
jlmer(r2 ~ Anger + Gender + (1 | id), lme4::VerbAgg, family = "binomial")
#> <Julia object of type GeneralizedLinearMixedModel>
#>
#> r2 ~ 1 + Anger + Gender + (1 | id)
#>
#> Variance components:
#> Column VarianceStd.Dev.
#> id (Intercept) 1.12074 1.05865
#> ────────────────────────────────────────────────────
#> Coef. Std. Error z Pr(>|z|)
#> ────────────────────────────────────────────────────
#> (Intercept) -1.10115 0.280681 -3.92 <1e-04
#> Anger 0.0462741 0.0134906 3.43 0.0006
#> Gender: M 0.260057 0.153847 1.69 0.0910
#> ────────────────────────────────────────────────────{broom}-style tidy() and
glance() methods for Julia regression models:
jmod <- jlmer(Reaction ~ Days + (Days | Subject), lme4::sleepstudy, REML = TRUE)
tidy(jmod)
#> effect group term estimate std.error statistic
#> 1 fixed <NA> (Intercept) 251.40510485 6.824597 36.838090
#> 2 fixed <NA> Days 10.46728596 1.545790 6.771481
#> 12 ran_pars Subject sd__(Intercept) 24.74065797 NA NA
#> 3 ran_pars Subject cor__(Intercept).Days 0.06555124 NA NA
#> 21 ran_pars Subject sd__Days 5.92213766 NA NA
#> 11 ran_pars Residual sd__Observation 25.59179572 NA NA
#> p.value
#> 1 4.537101e-297
#> 2 1.274703e-11
#> 12 NA
#> 3 NA
#> 21 NA
#> 11 NAglance(jmod)
#> nobs df sigma logLik AIC BIC deviance df.residual
#> 1 180 6 25.5918 NA NA NA 1743.628 174Experimental support for MixedModels.parametricbootstrap
via parametricbootstrap():
samp <- parametricbootstrap(jmod, nsim = 1000L, seed = 42L)
samp
#> <Julia object of type MixedModelBootstrap{Float64}>
#> MixedModelBootstrap with 1000 samples
#> parameter min q25 median mean q75 max
#> ┌───────────────────────────────────────────────────────────────────────────
#> 1 │ β1 227.464 246.884 251.608 251.655 256.229 275.687
#> 2 │ β2 4.99683 9.40303 10.4795 10.4522 11.5543 15.2264
#> 3 │ σ 21.0629 24.5779 25.5858 25.6024 26.5579 30.8176
#> 4 │ σ1 3.8862 19.8341 23.9517 23.8161 27.9909 40.7842
#> 5 │ σ2 1.65066 4.94152 5.77701 5.78757 6.61858 9.80011
#> 6 │ ρ1 -0.79257 -0.147053 0.0914976 0.111537 0.346284 1.0
#> 7 │ θ1 0.158198 0.762462 0.939845 0.935398 1.10182 1.72955
#> 8 │ θ2 -0.259593 -0.0358002 0.0185442 0.0197924 0.07347 0.333435
#> 9 │ θ3 0.0 0.175387 0.213763 0.207584 0.24721 0.402354tidy(samp)
#> effect group term estimate conf.low conf.high
#> 1 fixed <NA> (Intercept) 251.40510485 238.6573824 265.430084
#> 2 fixed <NA> Days 10.46728596 7.3121974 13.523381
#> 5 ran_pars Subject sd__(Intercept) 24.74065797 12.8464132 35.011690
#> 4 ran_pars Subject cor__(Intercept).Days 0.06555124 -0.4501055 1.000000
#> 6 ran_pars Subject sd__Days 5.92213766 3.3869360 8.310897
#> 3 ran_pars Residual sd__Observation 25.59179572 22.6637109 28.465550Check singular fit
issingular(jmod)
#> [1] FALSEList all properties of a MixedModel object (properties are accessible
via $)
propertynames(jmod)
#> [1] "A" "b" "beta" "betas" "corr" "dims"
#> [7] "feterm" "formula" "L" "lambda" "lowerbd" "objective"
#> [13] "optsum" "parmap" "PCA" "pvalues" "rePCA" "reterms"
#> [19] "sigma" "sigmarhos" "sigmas" "sqrtwts" "stderror" "theta"
#> [25] "u" "vcov" "X" "Xymat" "y" "β"
#> [31] "βs" "θ" "λ" "σ" "σs" "σρs"Example 1: extract PCA of random effects and return as an R list:
jmod$rePCA
#> <Julia object of type @NamedTuple{Subject::Vector{Float64}}>
#> (Subject = [0.5327756193675971, 1.0],)jl_get(jmod$rePCA)
#> $Subject
#> [1] 0.5327756 1.0000000Example 2: extract fitlog and plot
fitlog <- jl("refit!(x; thin=1)", x = jmod, .R = TRUE)$optsum$fitlog
thetas <- t(sapply(fitlog, `[[`, 1))
head(thetas)
#> [,1] [,2] [,3]
#> [1,] 1.00 0 1.00
#> [2,] 1.75 0 1.00
#> [3,] 1.00 1 1.00
#> [4,] 1.00 0 1.75
#> [5,] 0.25 0 1.00
#> [6,] 1.00 -1 1.00matplot(thetas, type = "o", xlab = "iterations")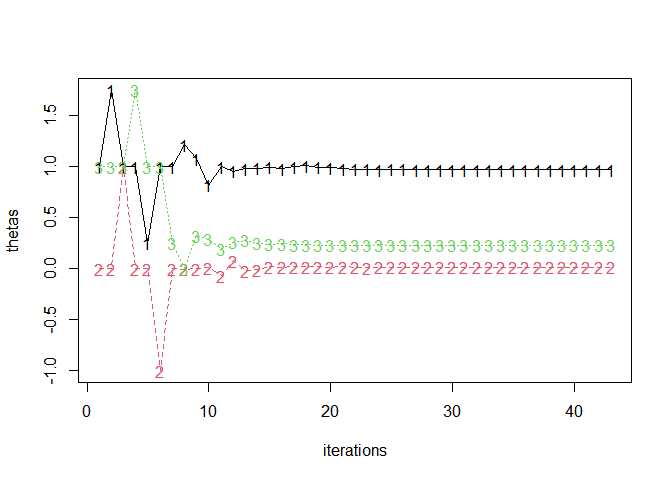
See information about the running Julia environment (e.g., the list
of loaded Julia libraries) with jlme_status():
jlme_status()
#> Julia Version 1.10.3
#> Commit 0b4590a550 (2024-04-30 10:59 UTC)
#> Build Info:
#> Official https://julialang.org/ release
#> Platform Info:
#> OS: Windows (x86_64-w64-mingw32)
#> CPU: 8 × 11th Gen Intel(R) Core(TM) i7-1165G7 @ 2.80GHz
#> WORD_SIZE: 64
#> LIBM: libopenlibm
#> LLVM: libLLVM-15.0.7 (ORCJIT, tigerlake)
#> Threads: 1 default, 0 interactive, 1 GC (on 8 virtual cores)
#>
#> Status `C:\Users\jchoe\AppData\Local\Temp\jl_UtfBLB\Project.toml`
#> [38e38edf] GLM v1.9.0
#> [98e50ef6] JuliaFormatter v1.0.60
#> [ff71e718] MixedModels v4.25.4
#> [3eaba693] StatsModels v0.7.4
#> [9a3f8284] Random{JuliaConnectoR}library(JuliaConnectoR)Use Julia(-esque) model-fitting syntax from R:
MixedModels <- juliaImport("MixedModels")
# Long-form construction of `jmod`
jmod2 <- MixedModels$fit(
MixedModels$LinearMixedModel,
juliaEval("@formula(Reaction ~ Days + (Days | Subject))"),
juliaPut(lme4::sleepstudy)
)
# In complete Julia syntax, using the sleepstudy dataset from MixedModels.jl
jmod3 <- juliaEval("
fit(
LinearMixedModel,
@formula(reaction ~ days + (days | subj)),
MixedModels.dataset(:sleepstudy)
)
")Be sure to pass integers to functions that expect Integer type,
(e.g., the MixedModels.parametricbootstrap() example
above):
jl_put(1)
#> <Julia object of type Float64>
#> 1.0jl_put(1L)
#> <Julia object of type Int64>
#> 1Using MKL.jl
or AppleAccelerate.jl
may improve model fitting performance (but see the system requirements
first).
# Not run
jlme_setup(add = "MKL", restart = TRUE)
jlme_status() # Should see MKL loaded hereIn practice, most of the overhead will come from transferring the
data from R to Julia. If you are looking to fit many models to the same
data, you should first filter to keep only used columns and then use
jl_data() to send the data to Julia. The Julia data frame
object can then be used to fit Julia models.
data_r <- mtcars
# Keep only columns you need + convert with `jl_data()`
data_julia <- jl_data(data_r[, c("mpg", "am")])
jlm(mpg ~ am, data_julia)
#> <Julia object of type StatsModels.TableRegressionModel>
#>
#> mpg ~ 1 + am
#>
#> ────────────────────────────────────────────────────────────────────────
#> Coef. Std. Error z Pr(>|z|) Lower 95% Upper 95%
#> ────────────────────────────────────────────────────────────────────────
#> (Intercept) 17.1474 1.1246 15.25 <1e-51 14.9432 19.3515
#> am 7.24494 1.76442 4.11 <1e-04 3.78674 10.7031
#> ────────────────────────────────────────────────────────────────────────If your data has custom contrasts, you can use
jl_contrasts() to also convert that to Julia first before
passing it to the model.
data_r$am <- as.factor(data_r$am)
contrasts(data_r$am) <- contr.sum(2)
data_julia <- jl_data(data_r[, c("mpg", "am")])
contrasts_julia <- jl_contrasts(data_r)
jlm(mpg ~ am, data_julia, contrasts = contrasts_julia)
#> <Julia object of type StatsModels.TableRegressionModel>
#>
#> mpg ~ 1 + am
#>
#> ────────────────────────────────────────────────────────────────────────
#> Coef. Std. Error z Pr(>|z|) Lower 95% Upper 95%
#> ────────────────────────────────────────────────────────────────────────
#> (Intercept) 20.7698 0.882211 23.54 <1e-99 19.0407 22.4989
#> am: 1 -3.62247 0.882211 -4.11 <1e-04 -5.35157 -1.89337
#> ────────────────────────────────────────────────────────────────────────If you spend non-negligible time fitting regression models for your work, please just learn Julia! It’s a great high-level language that feels close to R in syntax and its REPL-based workflow.
The JuliaConnectoR R package for powering the R interface to Julia.
The Julia packages GLM.jl and MixedModels.jl for fast implementations of (mixed effects) regression models.